Decoding Metabolism: From the subcellular to the single cell and whole-body level
Our Approach
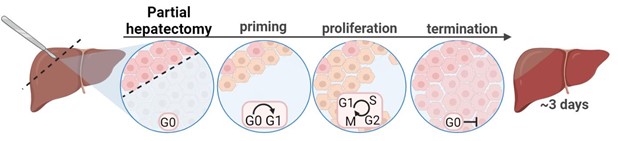
Our lab explores how metabolism functions across scales—from subcellular compartments to the whole organism level. To answer this, we use the liver’s extraordinary ability to regenerate as our model system. The liver is the only organ in the mammalian that can fully regenerate after significant injury or surgical resection, with the process in rodents completing in just a few days (Image 1). This unique regenerative capacity offers us a powerful window into the metabolic processes that drive cell division, growth, and tissue repair.
We combine in vivo physiology with advanced imaging technologies, classical molecular biology, and multi-omics techniques to capture metabolic dynamics across scales. By leveraging clinically relevant model systems, we aim to bridge the gap between basic science and a deeper understanding of both health and disease. We focus on multiple interlinked research questions, each exploring metabolism at a different level:
Subcellular Metabolism: The Metabolic Role of the Nucleus
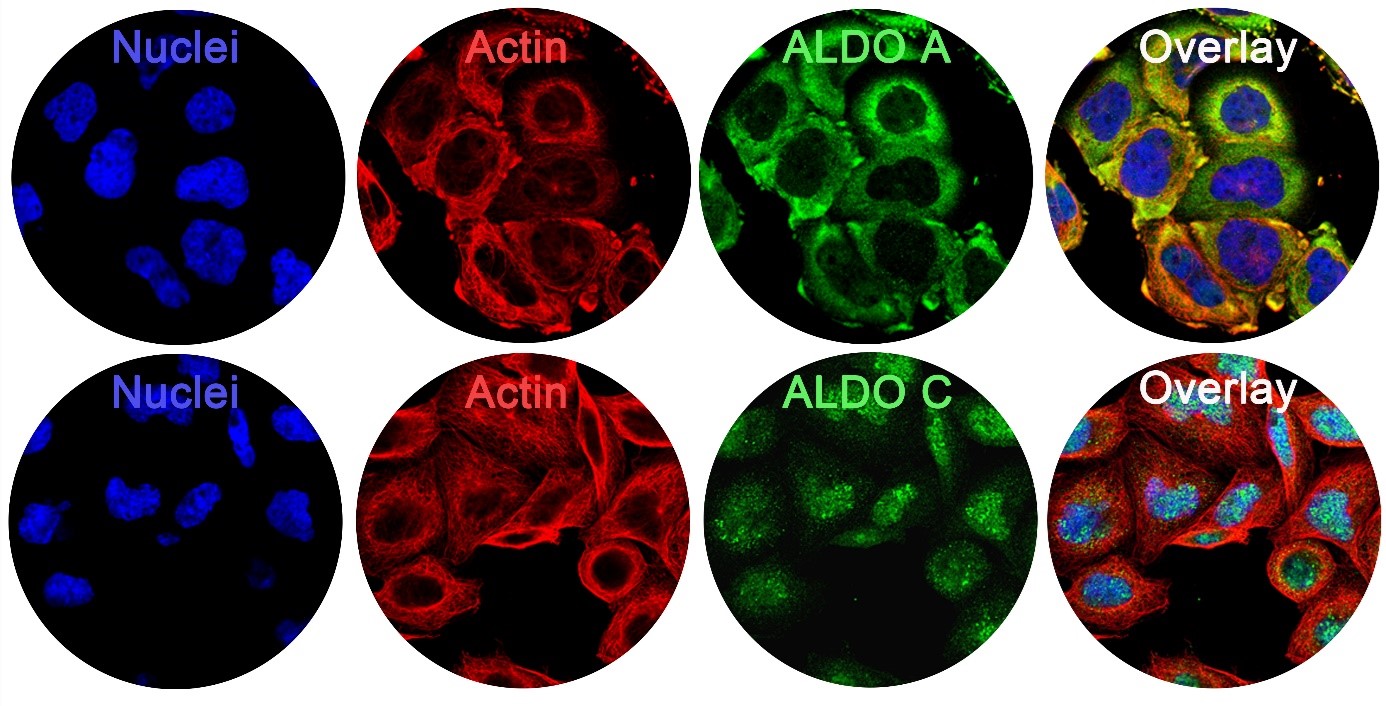
Cells have specialized compartments where metabolic reactions are precisely organized, ensuring efficiency and preventing harmful cross-reactions. However, the dynamic coordination of these metabolic pathways—especially in response to changing cellular needs—remains largely unexplored. Recent discoveries have shaken the classical view that glycolysis only occurs in the cytosol and the citric acid cycle happens solely in the mitochondria. We now know that selected metabolic enzymes can move into the cell's control center, the nucleus, to perform critical tasks (Image 2).
Our research seeks to unlock the mysteries of nuclear metabolism: which metabolic reactions occur within the nucleus and how they contribute to vital cellular processes like cell division, transcriptional regulation, and epigenetic modifications. Understanding nuclear metabolism could provide novel insights into how cells proliferate so efficiently in regeneration and how nuclear dysfunction could lead to diseases like cancer or liver fibrosis.
Single Cell Metabolism: Macrophages in Action
Macrophages are typically known for their role in immunity, fighting infections, and clearing dead cells. However, a more nuanced picture is emerging as macrophages are viewed as metabolic regulators that play a pivotal role in tissue repair. Our research focuses on macrophage metabolism, both inside individual cells and through their interactions with surrounding liver cells like hepatocytes.
Different macrophage types are coordinating between immune response and metabolic shifts to repair tissue efficiently. By exploring how intracellular metabolic pathways within specific macrophage subsets guide their behaviour, we aim to reveal how they maintain their energy balance while driving regeneration. Furthermore, macrophages communicate with surrounding cells, creating metabolic "crosstalk" that fine-tunes tissue repair (Image 3). Understanding these interactions helps us appreciate how the immune system shapes metabolic responses and opens up possibilities for interventions in liver diseases.
Whole-Body Metabolism: The Systemic View
The liver does not operate in isolation. As the body’s metabolic powerhouse, it must balance its regeneration with the energy demands of the entire organism (Image 4). Whole-body metabolism comes into play when resources are redirected to enable rapid liver growth. This is a remarkable biological task, where the body prioritizes liver repair without compromising other essential functions.

This part of our research dives into the systemic metabolic shifts that occur during liver regeneration. How do organs communicate during this process? How does the immune system, metabolic organs, and the nervous system collaborate to reallocate resources effectively? These questions are crucial to understanding regeneration in its full context—not just as a local process but as one that involves the entire body. This systemic view of metabolism offers exciting opportunities to explore how we might influence these processes in clinical settings. By understanding the global metabolic changes that support liver repair, we can identify key points where therapeutic interventions could promote regeneration and restore liver function.
- Miller A, York E, Stopka S, Martínez-François JR, Regan MS, Agar NYR, Yellen G. (2023) Nature Metabolism. Spatially resolved metabolomics and isotope tracing reveal dynamic metabolic responses of dentate granule neurons with acute stimulation.
- Miller A, Nagy C, Knapp B, Laengle J, Ponweiser E, Groeger M, Starkl P, Bergmann M, Wagner O, Haschemi A. (2017) Cell Metabolism. Exploring Metabolic Configurations of Single Cells within Complex Tissue Microenvironments.
- Kremslehner C, Miller A, Nica R, Nagelreiter I, Narz M, Golabia B, Vorstandlechner V, Mildnera M, Lachner J, Tschachler E, Ferraragh F, Klavins K, Schosserer M, Grillari J, Haschemi A, Gruber F. (2019) Redox Biology. Imaging of metabolic activity adaptations to UV stress, drugs and differentiation at cellular resolution in skin and skin equivalents
- Azzimato V, Chen P, Barreby E, Morgantini C, Levi L, Vankova A, Jager J, Sulen A, Diotallevi M, Shen J, Miller A, Ellis E, Rydén M, Näslund E, Thorell A, Lauschke V, Channon K, Crabtree M, Haschemi A, Craige S, Mori M, Spallotta F, Aouadi M. (2021) Gastroenterology. Hepatic miR-144 drives fumarase activity preventing NRF2 activation in obese livers.
- Anne Miller
I have always been fascinated by how metabolic reactions dynamically adapt through space and time driving my research to develop innovative solutions for long-standing scientific questions spanning fields from immunology to neuroscience. Just as I appreciate a slice of cake in the afternoon, I enjoy activities like reading, pilates, cycling, and traveling.
- Nyamdelger Sukhbaatar
I am a postdoc in the lab and my background is rooted in macrophage and immunological research, with a focus on how intracellular cues direct cell functions and phenotypes. I'm passionate about exploring how cells respond to their environment. Outside of the lab, I enjoy discovering new places and hiking in nature with my family
- Frédéric Soysouvanh
I am a postdoctoral researcher in the lab with a strong background in pathological wound healing involving tissue remodeling and fibrosis. My latest work on chronic liver disease convinced me to pursue my research on liver (patho)physiology. More recently, I have also explored immunological aspects during liver diseases. On my free time, I like cooking & baking, travelling, but only if it involves good food. I am a board games & puzzle enjoyer.
- Christina Hasler
As a master’s student, I am gaining valuable research experience and learning new molecular biology techniques. My curiosity drives me to explore the complex details of this field. In the long term, I aim to deepen this passion by pursuing a PhD. In my free time, I enjoy discovering new cities and spending time outdoors with my dog.